Physiochemical Characterization and Domain Annotation of ORF1ab Polyprotein of Novel Corona Virus 19 | SpringerLink

SIB on X: "“Whether you are a #PhDStudent looking for the ideal tool to analyse your #transcriptomics #data, or teaching undergraduates about #bioinformatics: #Expasy is the gateway to finding answers.” Christine Durinx,

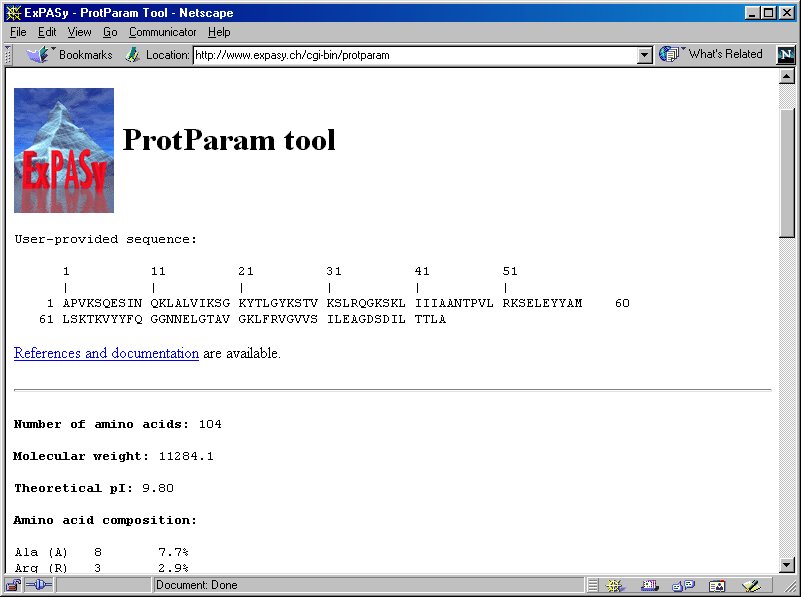
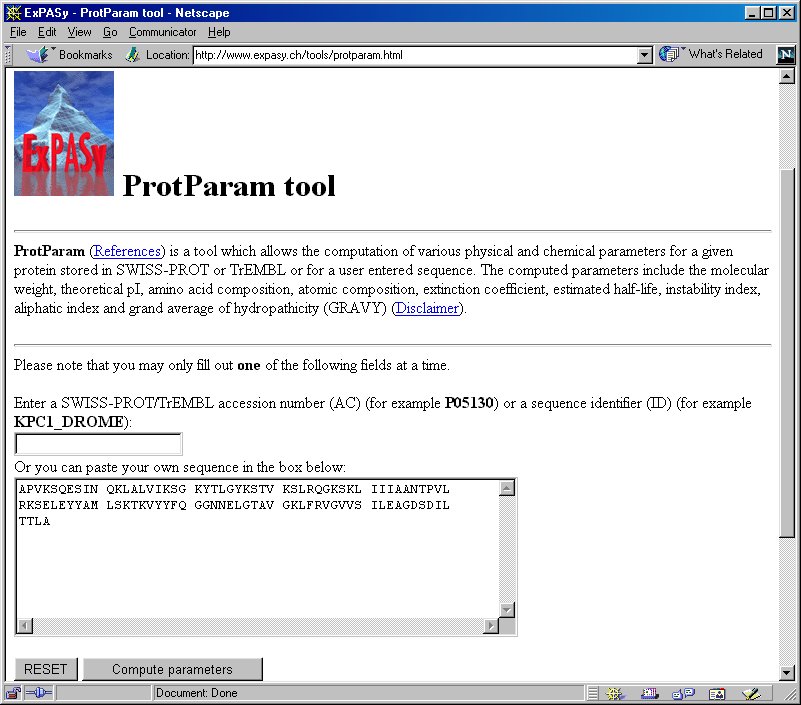
SOLVED: Use EXPASY Protparam tool to calculate the molecular weight, number of amino acids, and the theoretical pI, extinction coefficient under fully reducing conditions, and expected absorbance for 1 mg/ml protein. Use

SOLVED: Use EXPASY Protparam tool to calculate the molecular weight, number of amino acids, and the theoretical pI, extinction coefficient under fully reducing conditions, and expected absorbance for 1 mg/ml protein. Use